| Group | Genotype | Crossovers | Number | Totals |
| 1 | CShBz | None; the parentals | 479 | 952 |
| 2 | cshbz | 473 | ||
| 3 | C|shbz | Single; between C and others | 15 | 28 |
| 4 | c|ShBz | 13 | ||
| 5 | CSh|bz | Single, between Bz and others | 9 | 18 |
| 6 | csh|Bz | 9 | ||
| 7 | C|sh|Bz | Double recombinants | 1 | 2 |
| 8 | c|Sh|bz | 1 | ||
| Totals | 1000 |
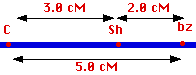
(But note that if you had simply been doing a testcross of a C,c,Sh,sh dihybrid, you would not have seen the double recombinants - they restored the parental linkage - so you would have understated the distance at 2.8 cM.)
(Again, a testcross between a Sh,sh,Bz,bz dihybrid would have only shown the 1.8% recombinants.)
And, in this case, a C,c,Bz,bz dihybrid would have missed the two recombination events because they restored the parental configuration, so the "true" value between the loci is the sum of the single recombinants plus twice the frequency of double recombinants.
 | Compare these results with the (less accurate) mapping results for the same loci done by test crossing dihybrids rather than trihybrids. |
A three-point cross also gives the gene order immediately.
The procedure is:| Welcome&Next Search |