BIOL 1400 -- Lecture Outline 18
"The best friend you'll ever have is a good head on your shoulders." -- Paraphrase
of an Icelandic saying
I. Genetic fingerprinting and PCR
- PCR, the polymerase chain reaction
- First unveiled in 1985, PCR is a technique for making
millions of copies of any given piece of DNA.
- It is possible to take an extremely tiny sample containing minute amounts of
DNA -- blood, hair, skin, etc. -- and "amplify" the DNA in it, making millions of
copies.
- How? When DNA replicates, the "work" is done by enzymes called DNA
polymerases.
- First, the DNA has to "unzip" -- in the lab, you can make this happen by heating
the DNA to just below the boiling point of water, causing it to denature and
break its hydrogen bonds. But that high a temperature would denature the DNA
polymerase that you would need to make new strands.
- Kary Mullis, working at Cetus Corp. in the San Francisco area, realized that you
could use the DNA polymerase from a heat-loving bacterium -- remember how in an
earlier lecture I mentioned that many bacteria can survive in very hot conditions?
They can do it 'cause they have unusually stable, denaturation-resistant enzymes. . .
- Using heat-resistant DNA polymerase enzymes, plus careful control of
temperature, Mullis developed a workable polymerase chain reaction -- and won
the Nobel Prize for his work in 1993.
- Click
here for a bit more background. . . .
- It is possible to use PCR to determine the sequence of bases in a piece
of DNA.
- This is done by creating millions of copies of the DNA, but ensuring that each
copy is stopped at a random nucleotide position, and then determining the last
nucleotide for each copy. . .
- In effect, what you find out is that, say. . . "Copies of this DNA 36 nucleotides long
end in A. . . copies 37 nucleotides long end in T. . . copies 38 nucleotides long end in
A. . . copies 39 nucleotides long end in G. . . copies 40 nucleotides long end in T. . . . .
copies 41 nucleotides long end in T. . . copies 42 nucelotides long end in C. . . "
and so on, until you have the whole sequence: . . . ATAGTTC. . .
- "Laid out" in order of size by a technique called gel electrophoresis (you've
seen this in lab), the fragments look like this:
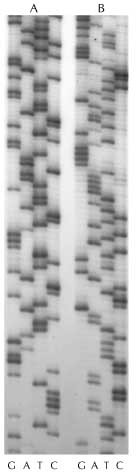
Results of a PCR analysis -- each column corresponds to one of the four
nucleotides
- This is what the fictitious scientists were doing in Jurassic Park,
incidentally. . .
- . . . and it's what's behind the famous Human Genome Project, a project
intended to sequence the complete human genome (a genome is the complete
set of genes of a living thing).
- Genetic fingerprinting
- One of the stranger discoveries of recent years: most of your DNA doesn't code
for proteins!
- Much of your DNA consists of long stretches of repeated nucleotides, such as
ACACACACACACACACACACACACACACACACACA. . . .
- This is sometimes called "junk DNA". . .
- . . . but it would be more accurate to say that we do not yet know what function,
if any, it has -- it may or may not be "junk".
- These long stretches are called RFLPs (if you want to get technical,
that's restriction fragment length polymorphisms). They vary in length
between individuals -- again, we're not sure why.
- Using PCR, we can determine the size of the RFLPs in a sample of DNA, and
then match a sample of blood, hair, semen, etc. to an individual.
- It's also possible to do paternity testing using RFLPs, since RFLPs are passed on
from parents to offspring. (Same principle as paternity testing using blood types,
but this is much more accurate in many cases.)
- PROBLEMS:
- PCR is very easy to contaminate! A small amount of foreign DNA -- say,
from a lab worker's dandruff -- can make it impossible to read a sample.
- Labs that do "fingerprinting" are subject to errors -- the PCR process itself
isn't always perfect, and other errors can creep in. Very careful quality
control is needed at every step -- and many crime labs have been caught doing
shoddy work.
- It's still not always certain just how unique a DNA fingerprint is, although
the more RFLPs you can look at, the greater the chance that the pattern is unique.
Go to Previous Notes |
Return to Lecture Schedule |
Return to Syllabus |
Contact the Prof |
Go to Next Notes
