What are Bacteria?
Until recently, the term bacteria was used for all microscopic prokaryotes. But, it turns out that there are two groups of prokaryotes that differ from each other in just about every way except size and lack of a nucleus.
These are now distinguished as the:
- Bacteria; the "true" bacteria (also known as Eubacteria)
- Archaea; (also known as Archaebacteria)
The archaea are so different from the bacteria that they must have had a long, independent evolutionary history since close to the dawn of life. In fact, there is considerable evidence that you are more closely related to the archaea than they are to the bacteria!
|
Index to this page
|
- prokaryotic (no membrane-enclosed nucleus)
- no mitochondria or chloroplasts
- a single chromosome
- a closed circle of double-stranded DNA
- with no associated histones
- If flagella are present, they are made of a single filament of the protein flagellin; there are none of the "9+2" tubulin-containing microtubules of the eukaryotes.
- ribosomes differ in their structure from those of eukaryotes
- have a rigid cell wall made of peptidoglycan.
- The plasma membrane is a phospholipid bilayer but contains no cholesterol or other steroids.
- no mitosis
- mostly asexual reproduction
- any sexual reproduction very different from that of eukaryotes; no meiosis
- Many bacteria form a single spore when their food supply runs low. Most of the water is removed from the spore and metabolism ceases. Spores are so resistant to adverse conditions of dryness and temperature that they may remain viable even after 50 years of dormancy.
Until recently classification has done on the basis of such traits as:
- shape
- bacilli: rod-shaped
- cocci: spherical
- spirilla: curved walls
- ability to form spores
- method of energy production (glycolysis for anaerobes, cellular respiration for aerobes
- nutritional requirements
- reaction to the Gram stain.
The Gram stain is named after the 19th century Danish bacteriologist who developed it.
- The bacterial cells are first stained with a purple dye called crystal violet.
- Then the preparation is treated with alcohol or acetone.
- This washes the stain out of gram-negative cells.
- To see them now requires the use of a counterstain of a different color (e.g., the pink of safranin).
- Bacteria that are not decolorized by the alcohol/acetone wash are gram-positive.
Although the Gram stain might seem an arbitrary criterion to use in bacterial taxonomy, it does, in fact, distinguish between two fundamentally different kinds of bacterial cell walls and reflects a natural division among the bacteria.
More recently, genome sequencing, especially of their 16S ribosomal RNA (rRNA), has provided new insights into the evolutionary relationships among the bacteria.
The Proteobacteria
This large group of bacteria form a clade sharing related rRNA sequences. They are all gram-negative but come in every shape (rods, cocci, spirilla).
They are further subdivided into 5 clades: alpha-, beta-, gamma-, delta-, and epsilon proteobacteria.
Some examples:
- Rickettsias.
These bacteria are too small to be clearly seen under the light microscope. Almost all are obligate intracellular parasites. This means that they can only grow and reproduce while within the living cells of their host — certain arthropods (ticks, mites, lice, fleas) and mammals.
- Rickettsia prowazekii causes typhus fever when it is transmitted to humans by lice.
- Rocky Mountain spotted fever is a rickettsial disease transmitted by ticks.
The mitochondria of eukaryotes probably evolved from endosymbiotic prokaryotes. Because of the similarities of their genomes, rickettsias may be the closest relatives to the ancestors of mitochondria.
- Rhizobia. These bacteria live in a mutualistic relationship with the roots of legumes where they are able to "fix" nitrogen (N2) in the air into compounds that can be used by living things.
- Magnetospirillum magnetotacticum
- Agrobacterium tumefaciens
- Sulfur bacteria.
Certain colorless bacteria share the ability of chlorophyll-containing organisms to manufacture carbohydrates from inorganic raw materials, but they do not use light energy for this. These so-called chemoautotrophic bacteria secure the necessary energy by oxidizing some reduced substance present in their environment. The free energy released by the oxidation is harnessed to the manufacture of food.
For example, some chemoautotrophic sulfur bacteria oxidize H2S in their surroundings (e.g., the water of sulfur springs) to produce energy:
2H2S + O2 → 2S + 2H2O; ΔG = -100 kcal
They then use this energy to reduce carbon dioxide to carbohydrate (like the photosynthetic purple sulfur bacteria).
2H2S + CO2 → (CH2O) + H2O + 2S
- Iron bacteria.
These chemoautotrophs are responsible for the brownish scale that forms inside the tanks of flush toilets. They complete the oxidation of partially oxidized iron compounds and are able to couple the energy produced to the synthesis of carbohydrate.
- Nitrosomonas
This chemoautotroph oxidizes NH3 (produced from proteins by decay bacteria) to nitrites (NO2-). This provides the energy to drive their anabolic reactions. The nitrites are then converted (by other nitrifying bacteria) into nitrates (NO3-), which supply the nitrogen needs of plants.
- Three important human pathogens in the β-proteobacteria.
.
The largest and most diverse subgroup of the proteobacteria.
Some examples
- Escherichia coli. The most thoroughly-studied of all creatures (possibly excepting ourselves). Its entire genome has been determined down to the last nucleotide: 4,639,221 base pairs of DNA encoding 4,377 genes. Lives in the human colon, usually harmlessly. However, water or undercooked food contaminated with the O157:H7 strain has caused severe — occasionally fatal — infections.
- Salmonella enterica. Two major human pathogens:
- Salmonella enterica var Typhi. Causes typhoid fever, a serious systemic infection occurring only in humans. This microbe is also known as Salmonella typhi.
- Salmonella enterica var Typhimurium. Confined to the intestine, it is a frequent cause of human gastrointestinal upsets but is also found in many other animals (that are often the source of the human infection). Also known as Salmonella typhimurium.
- Vibrio cholerae. Causes cholera, one of the most devastating of the intestinal diseases. The bacteria liberate a toxin that causes massive diarrhea (10–15 liters per day) and loss of salts. Unless the water and salts are replaced quickly, the victim may die (of shock) in a few hours. Like other intestinal diseases, cholera is contracted by ingestion of food or, more often, water that is contaminated with the bacteria.
- Pseudomonas aeruginosa. A common inhabitant of soil and water, it can cause serious illness in humans with
Frequently encountered in hospitals and resistant to most antibiotics and disinfectants.
- Yersinia pestis. This bacillus causes bubonic plague. It is usually transmitted to humans by the bite of an infected flea. As it spreads into the lymph nodes, it causes them to become greatly swollen, hence the name "bubonic" (bubo — swelling of a lymph node) plague. Once in the lungs, however, the bacteria can spread through the air causing the rapidly lethal (2–3 days) "pneumonic" plague. Untreated, 50–75 % of the cases of bubonic plague are fatal, and the figure for the pneumonic form reaches 100%.
The recurrent epidemics of the "black death" in Europe from 1347–1352, which killed off at least one quarter of the population, are thought by many (not all) to have been caused by this organism.
Although no major epidemics have occurred in this century, the threat is not entirely over. Yersinia pestis still flourishes in some rodent populations in the western U. S. and causes a few cases of human plague — primarily among small game hunters — each year (nine in 1998).
Completion of the DNA sequence of the genome of Y. pestis — its chromosome plus three plasmids — was reported on 4 October 2001.
- Francisella tularensis causes tularemia. This is primarily a disease of small mammals, but about 100 people become infected each year in the United States. Most cases occur in south-central states (KS, MO, OK, AR). However, the import of infected rabbits by game clubs has introduced the disease to Cape Cod and Martha's Vineyard in Massachusetts. In the summer of 2000, 15 people became ill (one died) on the island. All seem to have acquired their infection as they used lawn mowers and brush cutters that presumably stirred up the organism from the carcasses of infected animals.
- Haemophilus influenzae was once thought to cause influenza. It does not, but it can cause bacterial meningitis and middle ear infections in children and pneumonia in adults — especially those whose resistance is lowered by other diseases (e.g., AIDS). There is now an effective vaccine against the most dangerous strains. The complete genome of Haemophilus influenzae is known: 1,830,138 bp of DNA encoding 1,743 genes.
- Purple Sulfur Bacteria
Like green plants, these bacteria are photosynthetic, using the energy of sunlight to reduce carbon dioxide to carbohydrate. Unlike plants, however, they do not use water as a source of electrons.
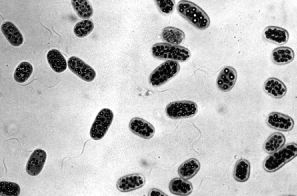
Instead they use hydrogen sulfide to supply the electrons needed to synthesize NADPH and ATP.
2H2S + CO2 → (CH2O) + H2O + 2S
In the process, they produce elemental sulfur (often — as seen in this photomicrograph of Chromatium — stored as granules within the cell). [Image from H. G. Schlegel and N. Pfennig, Arch. Microbiol. 38[1], 1961.]
Photosynthetic bacteria contain special types of chlorophylls (called bacteriochlorophylls) incorporated into membranes. With this machinery, they can run photosystem I but not photosystem II (which explains their inability to use water as a source of electrons).
Most photosynthetic bacteria are obligate anaerobes; they cannot tolerate free oxygen. Thus they are restricted to such habitats as the surface of sediments at the bottom of shallow ponds and estuaries. Here they must make do with whatever radiant energy gets through the green algae and aquatic plants growing above them. However, the absorption spectrum of their bacteriochlorophylls lies mostly in the infrared region of the spectrum so they can trap energy missed by the green plants above them.
This group contains the myxobacteria. They are found in vast numbers in soil and are major players in the decay of organic matter. [Link to discussion]
Two members of this small group that are human pathogens:
- Helicobacter pylori, the main cause of stomach ulcers [Link]
- Campylobacter jejuni; the bacterium most frequently implicated in gastrointestinal upsets. [Link]
Some Gram-Positive Bacteria
Aerobic Gram-Positive Rods
- Bacillus anthracis/cereus/thuringiensis. These organisms differ mainly in the plasmids they contain.
- B. anthracis causes anthrax. Currently the biological agent being used by terrorists. Its 2 plasmids contain the genes needed to synthesize
- a capsule which (like those of pneumococci) makes it resistant to phagocytosis, and
- the three components of the toxin that causes the disease symptoms. [More]
- B. thuringiensis — the organism, its toxin, and even the gene (also plasmid-encoded) for the toxin are used as biocontrol agents against a variety of insect pests. [Link to discussion]
- Bacillus subtilis. A common soil bacterium. Its chromosome contains 4,214,814 bp of DNA encoding 4,100 genes.
- Lactobacillus. Several species are used to convert milk into cheese, butter, and yogurt.
Anaerobic Gram-Positive Rods:
- Clostridium tetani. Clostridia are spore-forming obligate anaerobes. The spores of C. tetani are widespread in the soil and often get into the body through wounds. Puncture wounds (e.g., by splinters or nails) are particularly dangerous because they provide the anaerobic conditions needed for germination of the spores and growth of the bacteria.
C. tetani liberates a toxin that blocks transmitter release (by destroying the SNAREs needed) at inhibitory synapses in the spinal cord and brain. This interferes with the reciprocal inhibition of antagonistic pairs of skeletal muscles so the victim suffers violent muscle spasms. Fortunately, the disease — called tetanus — is now rare in developed countries, thanks to almost universal immunization against the toxin. Chemical alteration of the toxin produces a toxoid that still retains the epitopes of the toxin. Incorporated in a vaccine, the toxoid provides a relatively long-lasting (~10 years) immunity against tetanus.
- Clostridium botulinum. As little as 1 µg of its toxin eaten with an uncooked bean or mushroom can be fatal. The toxin blocks the release of acetylcholine (also by destroying the SNAREs needed for exocytosis) from the terminals of motor neurons. Thus the victim shows signs of sympathetic nervous activity (dilation of the pupils, inhibition of urination) and skeletal muscle weakness. If the intercostal muscles are effected, breathing may stop. The toxin is a protein and is quickly (10 minutes) denatured at 100 °C, so boiling home-canned products makes them safe to eat.
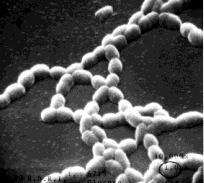 The bacteria in this group grow in characteristic colonies.
Most of these organisms grow as thin filaments — like a mold — rather than as single cells. In fact, they were long thought to be fungi. But fungi are eukaryotes and the actinomycetes are prokaryotes.
Actinomycetes dominate the microbial life in soil where they play a major role in the decay of dead organic matter. Many of them have turned out to be the source of valuable antibiotics, including streptomycin, erythromycin, and the tetracyclines.
These gram-positive organisms are closely related to the actinomycetes and often classified with them. They include three important human pathogens:
The bacteria in this group grow in characteristic colonies.
Most of these organisms grow as thin filaments — like a mold — rather than as single cells. In fact, they were long thought to be fungi. But fungi are eukaryotes and the actinomycetes are prokaryotes.
Actinomycetes dominate the microbial life in soil where they play a major role in the decay of dead organic matter. Many of them have turned out to be the source of valuable antibiotics, including streptomycin, erythromycin, and the tetracyclines.
These gram-positive organisms are closely related to the actinomycetes and often classified with them. They include three important human pathogens:
- Mycobacterium tuberculosis is the agent of tuberculosis (TB). Its genome contains 4,411,532 bp of DNA encoding some 3,959 genes.
- Mycobacterium leprae causes leprosy. Its genome contains 3,268,203 bp of DNA encoding only 1,604 genes.
Although a close relative of M. tuberculosis (they share 1,439 genes), much of its DNA encodes pseudogenes, genes that no longer make a functional product. M. leprae is an obligate intracellular parasite; it has never been cultured in vitro. This is probably because it has abandoned many of the genes needed for an independent existence choosing instead to depend on the genes of its host cell.
- Corynebacterium diphtheriae causes diphtheria. As in tetanus, it isn't the growth of the organism (in the throat) that is dangerous but the toxin it liberates. The toxin is the product of a latent bacteriophage in the bacterium. It catalyzes the inactivation of a factor necessary for amino acids to be added to the polypeptide chain being synthesized on the ribosome. Sensibly enough, the toxin has no such effect on the translation machinery of prokaryotes (or of chloroplasts and mitochondria).
Treatment of the toxin with formaldehyde converts it into a harmless toxoid. Immunization with this toxoid — usually incorporated along with tetanus toxoid and pertussis antigens in a "triple vaccine" (DTP) — protects against the disease.
These are thin, corkscrew-shaped, flexible organisms that range in length from a few to as many as 500 µm.
Two notorious examples:
- Treponema pallidum, the cause of syphilis, one of the most dangerous of the sexually transmitted diseases (STDs).
- Borrelia burgdorferi is transmitted to humans through the bite of a deer tick causing Lyme disease (16,801 cases were reported in the U.S. in 1998).
Both these organisms have had their complete genomes sequenced. [Link]
Mycoplasmas have the distinction of being the smallest living organisms. They are so small (0.1 µm) that they can be seen only under the electron microscope.
Mycoplasmas are obligate parasites; that is, they can live only within the cells of other organisms. They are probably the descendants of gram-positive bacteria who have lost much of their genome — now depending on the gene products of their host.
The DNA sequences of the complete genomes of seven mycoplasmas have been determined, including
- Mycoplasma genitalium has 580,073 base pairs of DNA encoding 517 genes (480 for proteins; the rest for RNAs).
- Mycoplasma urealyticum has 751,719 base pairs of DNA encoding 651 genes (613 for proteins; 39 for RNAs).
- Mycoplasma pneumoniae has 816,394 base pairs of DNA encoding 679 genes.
The scientists at The Institute for Genomic Research (TIGR) who determined the Mycoplasma genitalium sequence followed this work by systematically destroying its genes (by mutating them with insertions) to see which ones are essential to life and which are dispensable. Of the 480 protein-encoding genes, they conclude that only 265–350 of them are essential to life.
Chlamydiae are also obligate intracellular parasites.
- Chlamydia trachomatis
Its genome contains 1,042,519 bp of DNA encoding 938 genes.
In 1998, this organism infected 604,420 people in the U. S. The infection is usually spread by sexual intercourse making it the most common sexually-transmitted disease (STD). It is easily cured if diagnosed, but many infections remain untreated and, in females, are a major cause of pelvic inflammatory disease. This causes scarring of the uterus and fallopian tubes and often results in infertility.
Mothers can pass the infection on to their newborn babies causing serious eye disease and pneumonia. To avoid this, pregnant women are usually tested for chlamydia and treated with antibiotics if they are infected.
Several million people in the desert regions of Asia, Africa, and the Near East have been blinded by trachoma. This eye infection is caused by a strain of C. trachomatis (and is responsible for its name).
- Chlamydia psittaci usually infects birds, but can infect their human contacts causing psittacosis (a.k.a. ornithosis).
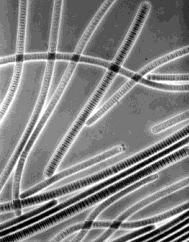 Unlike other photosynthetic prokaryotes, cyanobacteria
Cyanobacteria also contain two antenna pigments:
Unlike other photosynthetic prokaryotes, cyanobacteria
Cyanobacteria also contain two antenna pigments:
- blue phycocyanin (making them "blue-green") and
- red phycoerythrin (The Red Sea gets its name from the periodic blooms of red-colored cyanobacteria.)
These two pigments also occur in red algae. Perhaps their chloroplasts evolved from endosymbiotic cyanobacteria. In fact, probably all chloroplasts evolved from endosymbiotic cyanobacteria.
The micrograph is of Oscillatoria, a filamentous cyanobacterium (magnified about 800 times). Each disk in the chains is one cell.
There is now lots of evidence that both of these eukaryotic organelles evolved from once free-living bacteria.
- alpha-proteobacteria like the rickettsias for mitochondria
- cyanobacteria for chloroplasts
that took up an endosymbiotic way of life in the ancestors of the eukaryotes.
19 December 2004

 The bacteria in this group grow in characteristic colonies.
The bacteria in this group grow in characteristic colonies.
 Unlike other photosynthetic prokaryotes, cyanobacteria
Unlike other photosynthetic prokaryotes, cyanobacteria