| Index to this page |
The nucleus is enveloped by a pair of membranes enclosing a lumen that is continuous with that of the endoplasmic reticulum. The inner membrane is stabilized by a meshwork of intermediate filament proteins called lamins.
The nuclear envelope is perforated by thousands of nuclear pore complexes (NPCs) that control the passage of molecules in and out of the nucleus.
Most of the protein consists of multiple copies of 5 kinds of histones. These are basic proteins, bristling with positively charged arginine and lysine residues. (Both Arg and Lys have a free amino group on their R group, which attracts protons (H+) giving them a positive charge.) Just the choice of amino acids you would make to bind tightly to the negatively-charged phosphate groups of DNA.
Chromatin also contains small amounts of a wide variety of nonhistone proteins. Most of these are transcription factors (e.g., the steroid receptors) and their association with the DNA is more transient.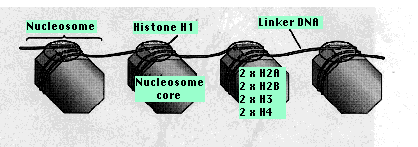 Two copies of each of four kinds of histones
Two copies of each of four kinds of histones
From 20–60 bp of DNA link one nucleosome to the next. Each linker region is occupied by a single molecule of histone 1 (H1).
The binding of histones to DNA does not depend on particular nucleotide sequences in the DNA but does depend critically on the amino acid sequence of the histone. Histones are some of the most conserved molecules during the course of evolution. Histone H4 in the calf differs from H4 in the pea plant at only 2 amino acids residues in the chain of 102.
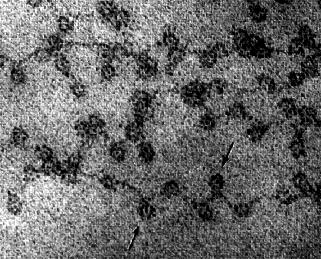
This electron micrograph (courtesy of David E. Olins and Ada L. Olins) shows chromatin from the nucleus of a chicken red blood cell (birds, unlike most mammals, retain the nucleus in their mature red blood cells). The arrows point to the nucleosomes. You can see why the arrangement of nucleosomes has been likened to "beads on a string".
The formation of nucleosomes helps somewhat, but not nearly enough, to make the DNA sufficiently compact to fit in the nucleus. In order to fit 46 DNA molecules (in humans), totaling over 2 meters in length, into a nucleus that may be only 10 µm across requires more extensive folding and compaction.
Although their amino acid sequence (primary structure) is unvarying, individual histone molecules do vary in structure as a result of chemical modifications that occur later to individual amino acids.
These include adding:The chemical modifications occur on these tails, especially of H3 and H4. Most of theses changes are reversible. For example, acetyl groups are
But there is surely more to the story.
In general, the "standard" histones are incorporated into the nucleosomes as new DNA is synthesized during S phase of the cell cycle. Later, some are replaced by variant histones as conditions in the cell dictate.
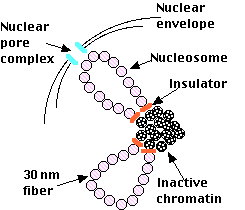 Euchromatin
Euchromatin
| More on insulators |
The diagram represents a hypothetical model of how euchromatin and heterochromatin may be organized during interphase in a vertebrate cell.
Transcription factors cannot bind to their promoter if the promoter is blocked by a nucleosome. For at least some genes, one of the first functions of the assembling transcription factors is to slide the nucleosome father along the DNA molecule exposing the gene's promoter so that the transcription factors can then bind to it.
The actual transcription of protein-coding genes is done by RNA polymerase II (RNAP II). In order for it to travel along the DNA to be transcribed, a complex of proteins removes the nucleosomes in front of it and then replaces them after RNAP II has transcribed that portion of DNA and moved on.
During the period between cell divisions, when the chromosomes are in their extended state, 1 or more of them (10 in human cells) have loops extending into a spherical mass called the nucleolus. Here are synthesized three (of the four) kinds of RNA molecules (28S, 18S, 5.8S) used in the assembly of the large and small subunits of ribosomes.
28S, 18S, and 5.8S ribosomal RNA is transcribed (by RNA polymerase I) from hundreds to thousands of tandemly-arranged rDNA genes distributed (in humans) on 10 different chromosomes. The rDNA-containing regions of these 10 chromosomes cluster together in the nucleolus.
(In yeast, the 5S rRNA molecules — as well as transfer RNA molecules — are also synthesized (by RNA polymerase III) in the nucleolus.)Once formed, rRNA molecules associate with the dozens of different ribosomal proteins used in the assembly of the large and small subunits of the ribosome.
But all proteins are synthesized in the cytosol — and all the ribosomes are needed in the cytosol to do their work — so there must be a mechanism for the transport of these large structures in and out of the nucleus. This is one of the functions of the nuclear pore complexes.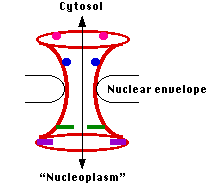
The nuclear envelope is perforated with thousands of pores.
Each is constructed from a number (30 in yeast; probably around 50 in vertebrates) different proteins called nucleoporins.The entire assembly forms an aqueous channel connecting the cytosol with the interior of the nucleus ("nucleoplasm"). When materials are to be transported through the pore, it opens up to form a channel some 25 nm wide — large enough to get such large assemblies as ribosomal subunits through.
Transport through the nuclear pore complexes is active; that is, it requires
All proteins are synthesized in the cytosol and those needed by the nucleus must be imported into it through the NPCs. Probably each of these proteins has a characteristic sequence of amino acids — called a nuclear localization sequence (NLS) — that targets it for entry.
They include:Both the RNA and protein molecules contain a characteristic nuclear export sequence (NES) needed to ensure their association with the right carrier molecules to take them out to the cytosol.
| Welcome&Next Search |