
| Index to this page |
Messenger RNA (mRNA) is single-stranded. Its sequence of nucleotides is called "sense" because it results in a gene product (protein). Normally, its unpaired nucleotides are "read" by transfer RNA anticodons as the ribosome proceeds to translate the message. (See mechanism of translation.)
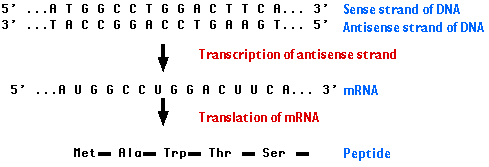
However, RNA can form duplexes just as DNA does. All that is needed is a second strand of RNA whose sequence of bases is complementary to the first strand; e.g.,
5´ C A U G 3´ mRNAThe second strand is called the antisense strand because its sequence of nucleotides is the complement of message sense. When mRNA forms a duplex with a complementary antisense RNA sequence, translation is blocked.
3´ G U A C 5´ Antisense RNA
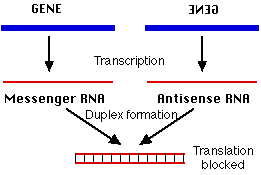 This may occur because
This may occur because
With recombinant DNA methods, synthetic genes (DNA) encoding antisense RNA molecules can be introduced into the organism.
Most tomatoes that have to be shipped to market are harvested before they are ripe. Otherwise, ethylene synthesized by the tomato causes them to ripen and spoil before they reach the customer.
Transgenic tomatoes have been constructed that carry in their genome an artificial gene (DNA) that is transcribed into an antisense RNA complementary to the mRNA for an enzyme involved in ethylene production. These tomatoes make only 10% of the normal amount of the enzyme.
The goal of this work was to provide supermarket tomatoes with something closer to the appearance and taste of tomatoes harvested when ripe. However, these tomatoes often became damaged during shipment and handling and have been taken off the market.
| Right: Flower of a tobacco plant carrying a transgene whose transcript is antisense to one of the mRNAs needed for normal flower pigmentation. Left: Flower of another transgenic plant that failed to have its normal pigmentation altered. (Courtesy of van der Krol, et. al., from Nature 333:866, 1988.) |  |
In this respect, it is easier to produce transgenic plants than transgenic animals.
Do cells contain genes that are naturally translated into antisense RNA molecules capable of blocking the translation of other genes in the cell? Recently a few cases have been found, and these seem to represent another method of regulating gene expression.
In both mice and humans, the gene for the insulin-like growth factor 2 receptor (Igf2r) that is inherited from the father synthesizes an antisense RNA that appears to block synthesis of the mRNA for Igf2r. An inherited difference in the expression of a gene depending on whether it is inherited from the mother or the father is called genomic or parental imprinting.
| Imprinting of the Igf2r gene. |
In testing the effects of antisense RNA, one should use sense RNA of the same coding region as a control. Surprisingly, preparations of sense RNA often turn out to be as effective an inhibitor as antisense RNA.
Why? It seems that the preparations of sense RNA often are contaminated with hybrids: sense and antisense strands that form a double helix of double-stranded RNA (dsRNA). Double-stranded RNA corresponding to a particular gene is a powerful suppressant of that gene. In fact, the suppressive effect of antisense RNA probably also depends on its ability to form dsRNA (using the corresponding mRNA as a template).
The ability of dsRNA to suppress the expression of a gene corresponding to its own sequence is called RNA interference (RNAi). It is also called post-transcriptional gene silencing or PTGS.
The only RNA molecules normally found in the cytoplasm of a cell are molecules of single-stranded RNA. If the cell finds molecules of double-stranded RNA (dsRNA), it uses an enzyme called Dicer to cut them into fragments containing 19 base pairs (~2 turns of a double helix) with two additional nucleotides at the opposite end of each strand.
The two strands of each fragment then separate — releasing the antisense strand. With the aid of a protein, it binds to a complementary sense sequence on a molecule of mRNA. If the base-pairing is exact, the mRNA is destroyed.
Because of their action, these fragments of RNA have been named "short (or small) interfering RNA" (siRNA).
The complex of siRNA and protein is called the "RNA-induced silencing complex" (RISC).
There is also evidence — in a variety of eukaryotes — that siRNAs can inhibit transcription of genes
How these siRNAs — synthesized in the cytosol — gain access to the DNA in the nucleus is unknown.
Synthetic siRNA molecules that bind to gene promoters can — in the laboratory — repress transcription of that gene. The repression is mediated by methylation of the DNA in the promoter and, perhaps, methylation of histones in the vicinity.
There is a strain of rice (LGC-1) that produces abnormally low levels of proteins called glutelins. It turns out that
RNAi has been found to operate in such diverse organisms as plants, fungi, and animals such as Drosophila melanogaster, Caenorhabditis elegans, and even mice and the zebrafish. Such a universal cell response must have an important function. What could it be?
Some possibilities:
In any case, the discovery of RNAi adds a promising tool to the toolbox of molecular biologists. Introducing dsRNA corresponding to a particular gene will knock out the cell's own expression of that gene. (Feeding C. elegans on E. coli manufacturing the dsRNA will even do the trick.)
Heroic Example
In the 24 March 2005 issue of Nature, Sönnichsen et al reported that they have injected dsRNAs corresponding to 20,326 of C. elegans's genes (98% of the total!) and monitored the effect of each on embryonic development from the completion of meiosis (following fertilization) through the second mitotic division that produces the 4-cell embryo.
They found that at least 661 different genes altered some process during this period:(Another thousand genes produced phenotypic effects that were seen at later stages of development.)
Because RNAi can be done in particular tissues at a chosen time, it often provides an advantage over conventional gene "knockouts" where the missing gene is carried in the germline and thus whose absence may kill the embryo before it can be studied.
| Link to discussion of knockout mice. |
The disadvantage to simply introducing dsRNA fragments into a cell is that gene expression is only temporarily reduced. However, Brummelkamp et. al. report in the 19 April 2002 issue of Science that they have succeeded in introducing into (mammalian) cells a DNA vector that can continuously synthesize a siRNA corresponding to the gene that they want to suppress. Two months later the cells still failed to manufacture the protein whose gene had been turned off by RNAi.
Two papers in the 25 July 2002 issue of Nature report that human cells in culture can be protected from by siRNAs with sequences that match RNA molecules encoded by the viruses.And the 19 June 2003 issue of Nature reports on coffee plants that were engineered to express a transgene that makes siRNA that interferes — by RNAi — with the expression of a gene needed to make caffeine. So perhaps "decaf" coffee will one day no longer require the chemical removal of caffeine from coffee beans.
Because its target is so specific, the possibility of using RNAi to shut down the expression of a single gene has created great excitement that a new class of therapeutic agents is on the horizon.
Antisense RNA that is complementary to the proto-oncogene BCL-2 is being examined as a possible therapy for certain B-cell lymphomas and leukemias.
But getting the antisense RNA into the correct cells is a difficult problem. One approach is to chemically modify the siRNA to enable it to enter cells. In mice, an intravenous injection of a chemically-modified siRNA specific for the mRNA of apolipoprotein B
| Another approach: Antisense oligodeoxynucleotides (ODNs) are synthetic molecules that — because they, too, are antisense — also block mRNA translation. One has been approved for human therapy. [Link] |
In C. elegans, successful development through its larval stages and on to the adult requires the presence of at least two "microRNAs" ("miRNAs") — single-stranded RNA molecules containing about 22 nucleotides and thus about the same size as siRNAs.
These small single-stranded transcripts are generated by the cleavage of larger precursors using the C. elegans version of Dicer.
They act by either destroying or inhibiting translation of several messenger RNAs in the worm (by binding to a region of complementary sequence in the 3' untranslated region [3'-UTR] of the mRNA).
The microRNAs (miRNAs) in C. elegans (which were first called "small temporal RNAs") turn out to be representatives of a large class of RNAs that
While direct evidence of the function of many of these newly-discovered gene products remains to be discovered, they probably regulate gene expression by regulating messenger RNA (mRNA), either
| Link to a discussion of riboswitches — another mechanism for regulating the translation of mRNA. |
These "riboregulators" have two traits ideally suited for this:
A study reported in Cell (Rhoades, et al., 110: 513, 23 Aug 2002) found that most of the miRNAs they examined in Arabidopsis matched the messenger RNAs (mRNAs) of transcription factors. Presumably these miRNAs repress their target mRNA. If so, this could provide a mechanism for daughter cells to escape from the transcription factors of their parent cell and thus be able to enter a different path of differentiation.
Of the 46 miRNAs expressed in the Drosophila embryo, 25 have been shown to be essential to normal development. (See Leaman, D., et al., Cell, 1 July 2005.)
MicroRNAs regulate (repress) expression of genes in mammals as well. Genome analysis has revealed thousands of human genes whose transcripts (mRNAs) contain sequences to which one or more of our miRNAs might bind. RAS, for example, has binding sites for a miRNA, and a defect in the production of that miRNA is associated with especially aggressive lung cancer.
And a recent study reported in Nature (Lim, et al., 433: 769, 17 Feb 2005) used DNA chip analysis to show that when a particular miRNA was expressed in HeLa cells,This repression of gene expression (their protein products) by miRNAs is surely more than simply a laboratory curiosity. In mice, overexpression of a particular miRNA during embryonic development prevents the heart from developing normally and leads to the early death of the mouse.
So miRNAs may play as important role as transcription factors in coordinating the expression of multiple genes in a particular type of cell at particular times.
In addition to protein transcription factors, eukaryotes use small RNA molecules to regulate gene expression — almost always by repressing it — so the phenomenon is called RNA silencing.
There are two sources of small RNA molecules:
Aside from their use as laboratory — and perhaps therapeutic — tools, small RNAs are clearly essential to the organisms that make them.
Some examples:
| Welcome&Next Search |